A Review of Graph Neural Networks in Epidemic Modeling
Time and Location
Time: Sunday, August 25 during 10 am - 1 pm (Barcelona Time)
Zoom Link: Please use the link on KDD virtual platform
Abstract
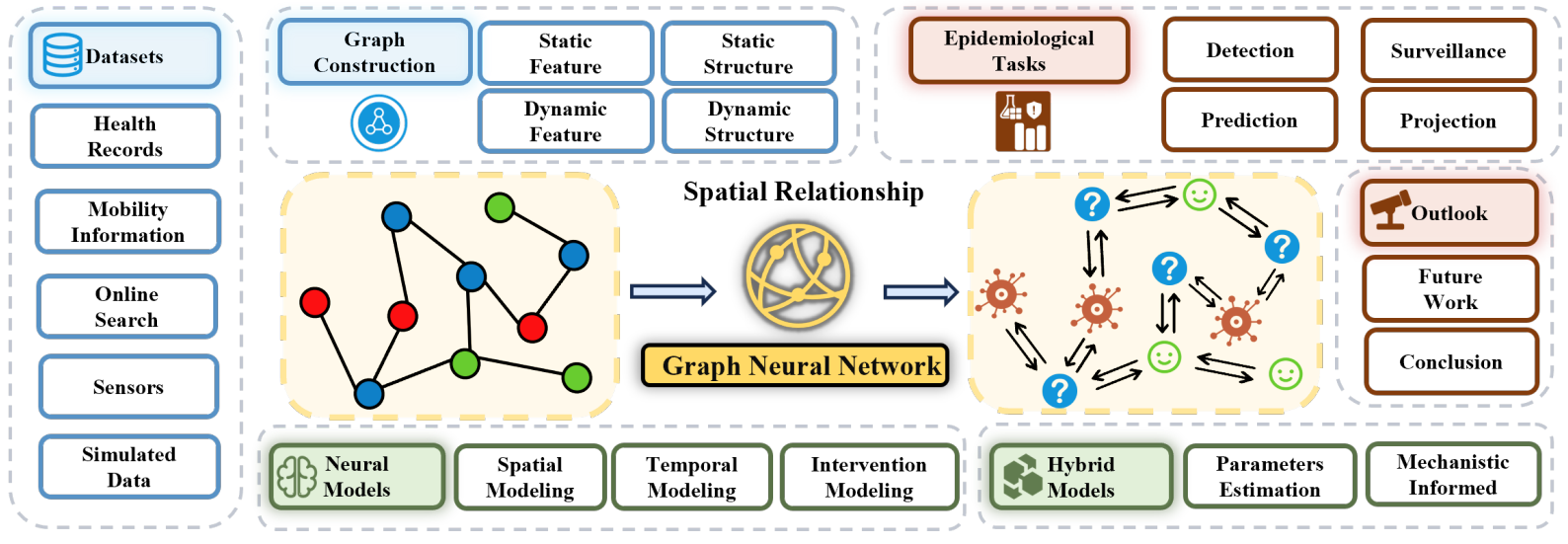
Since the onset of the COVID-19 pandemic, there has been a growing interest in studying epidemiological models. Traditional mechanistic models mathematically describe the transmission mechanisms of infectious diseases. However, they often fall short when confronted with the growing challenges of today. Consequently, Graph Neural Networks (GNNs) have emerged as a progressively popular tool in epidemic research. In this paper, we endeavor to furnish a comprehensive review of GNNs in epidemic tasks and highlight potential future directions. To accomplish this objective, we introduce hierarchical taxonomies for both epidemic tasks and methodologies, offering a trajectory of development within this domain. For epidemic tasks, we establish a taxonomy akin to those typically employed within the epidemic domain. For methodology, we categorize existing work into Neural Models and Hybrid Models. Following this, we perform an exhaustive and systematic examination of the methodologies, encompassing both the tasks and their technical details. Furthermore, we discuss the limitations of existing methods from diverse perspectives and systematically propose future research directions. We envisage that this tutorial will serve as a valuable resource for researchers, delivering a thorough examination of contemporary GNN models in epidemiology, while also illuminating promising avenues for future exploration.
Target Audience
This tutorial is designed for researchers and practitioners from both the graph machine learning and epidemiology communities, who are interested in exploring the application of GNNs in epidemiology. It is recommended that participants possess a foundational knowledge of (un)supervised learning, neural networks, and basic epidemiological concepts. Furthermore, prior experience in graph learning (eg., training a GNN) will be beneficial for attendees.
Tutorial Syllabus
The topics of this full-day tutorial include (but are not limited to) the following:
Graph theory and Graph Fourier Analysis
Foundations of Graph Neural Networks
Mechanistic Models
Hybrid Models in Epidemiology
EpiLearn package for Epidemic Modeling
The tutorial outline is shown below:
* Introduction and Background (20 minutes)
Epidemic Modeling
Graph Machine Learning and Graph Neural Networks
* Comprehensive Taxonomies (40 minutes)
Epidemiological Tasks
Epidemiological Datasets
Graph Construction
* Break (15 minutes)
* Exploring Neural and Hybrid Models (60 minutes)
Methodological Traits
Neural Models
Hybrid Models
* Break (15 minutes)
* EpiLearn: A Python Library for Deep Learning in Epidemic Data Modeling and Analysis (10 minutes)
* Future Directions and Conclusions (20 minutes)
Epidemic at Scales
Cross-Modality in Epidemiology
Epidemic Diffusion Processes
Interventions for Epidemics
Generating Explainable Predictions
Addressing Challenges in Epidemic Data
Tutorial slides
Opensource Project
EpiLearn is a Pytorch-based machine learning tool-kit for epidemic data modeling and analysis. We provide numerous features including:
- Implementation of Epidemic Models
- Simulation of Epidemic Spreading
- Visualization of Epidemic Data
- Unified Pipeline for Epidemic Tasks
For more details, please refer to: EpiLearn
Presenters

Zewen Liu
Emory University, USA

Guancheng Wan
Emory University, USA

B. Aditya Prakash
Georgia Institute of Technology, USA

Max S. Y. Lau
Emory University, USA

Wei Jin
Emory University, USA